
The driving goal of my laboratory is to understand the regulatory roles and biochemical mechanisms of RNA modifying enzymes and sex-specific RNA regulation in human disease. It has been demonstrated recently that a diverse set of enzyme-mediated modifications are found internally within RNAs, which markedly influence the fate of RNAs in cells. Many enzymes responsible for regulating protein and DNA modifications are targets of current therapies. RNA epitranscriptomics, the study of RNA modifications, is the new frontier in this arena. However, there are many fundamentally important questions, such as whether RNA modifications synergistically impact gene regulation, a new research area that my lab spearheaded in the RNA modification field. Our work studies the coordination of modification across RNA species through (1) interactions of RNA modifying enzymes (indirect model) and direct interactions of mRNA, tRNA, and rRNA modifications during translation.
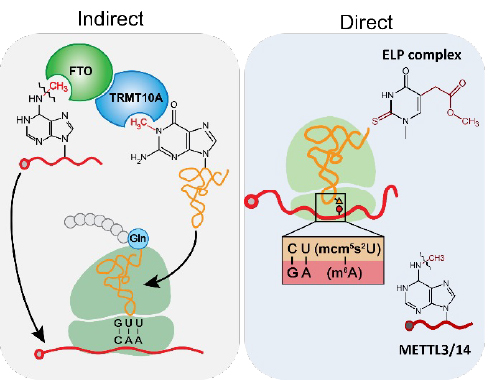
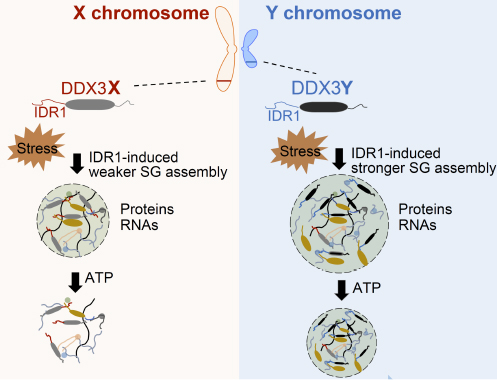
Furthermore, based on our observation of sex-specific RNA binding proteins, we have been investigating how sex and gender influence gene regulation at the RNA level. Sex differences are evident in human diseases. One of the significant keys to sex-biased differences lies in the sex chromosomes. Our work focuses on the functions of Y chromosome-encoded RNA binding proteins in non-reproductive organs. And the functional differences between Y chromosome-encoded RNA binding proteins and their homologous proteins on the X chromosomes.

